A | B | C | D | E | F | G | H | CH | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
Inositol monophosphatase 1 is an enzyme that in humans is encoded by the IMPA1 gene.[5][6]
Interacting partners
IMPA1 has been shown to interact with Bergmann glial S100B[7] and calbindin.[8][9]
Chemical inhibitors
L-690,330 is a competitive inhibitor of IMPase activity with very good activity in vitro however with limited bioavailability in vivo.[10] Due to its increased specificity compared to Lithium, L-690,330 has been used extensively in characterizing the results of IMPase inhibition in various cell culture models. L-690,488, a prodrug or L-690,330, has also been developed which has greater cell permeability. Treatment of cortical slices with L-690,488 resulted in accumulation of inositol demonstrating the activity of this inhibitor in tissue.[11]
Inhibition of IMPA1 activity can have pleiotropic effects on cellular function, including altering phosphoinositide signalling,[12] autophagy, apoptosis,[13] and other effects.
Bipolar disorder
Initially it was noticed that several drugs useful in treatment of bipolar disorder such as lithium, carbamazepine and valproic acid had a common mechanism of action on enzymes in the phosphatidylinositol signalling pathway[14] and the inositol depletion hypothesis for the pathophysiology of bipolar disorder was suggested. Intensive research has so far not confirmed this hypothesis, partly because lithium can also act on a number of other enzymes in this pathway, complicating results from in vitro studies.
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000133731 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000027531 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ McAllister G, Whiting P, Hammond EA, Knowles MR, Atack JR, Bailey FJ, Maigetter R, Ragan CI (Aug 1992). "cDNA cloning of human and rat brain myo-inositol monophosphatase. Expression and characterization of the human recombinant enzyme". Biochem J. 284 (3): 749–54. doi:10.1042/bj2840749. PMC 1132602. PMID 1377913.
- ^ "Entrez Gene: IMPA1 inositol(myo)-1(or 4)-monophosphatase 1".
- ^ Vig PJ, Shao Q, Subramony SH, Lopez ME, Safaya E (September 2009). "Bergmann glial S100B activates myo-inositol monophosphatase 1 and Co-localizes to purkinje cell vacuoles in SCA1 transgenic mice". Cerebellum. 8 (3): 231–44. doi:10.1007/s12311-009-0125-5. PMC 3351107. PMID 19593677.
- ^ Schmidt H, Schwaller B, Eilers J (April 2005). "Calbindin D28k targets myo-inositol monophosphatase in spines and dendrites of cerebellar Purkinje neurons". Proc. Natl. Acad. Sci. U.S.A. 102 (16): 5850–5. Bibcode:2005PNAS..102.5850S. doi:10.1073/pnas.0407855102. PMC 556286. PMID 15809430.
- ^ Berggard T, Szczepankiewicz O, Thulin E, Linse S (November 2002). "Myo-inositol monophosphatase is an activated target of calbindin D28k". J. Biol. Chem. 277 (44): 41954–9. doi:10.1074/jbc.M203492200. PMID 12176979.
- ^ Atack JR, Cook SM, Watt AP, Fletcher SR, Ragan CI (February 1993). "In vitro and in vivo inhibition of inositol monophosphatase by the bisphosphonate L-690,330". J. Neurochem. 60 (2): 652–8. doi:10.1111/j.1471-4159.1993.tb03197.x. PMID 8380439. S2CID 23498954.
- ^ Atack JR, Prior AM, Fletcher SR, Quirk K, McKernan R, Ragan CI (July 1994). "Effects of L-690,488, a prodrug of the bisphosphonate inositol monophosphatase inhibitor L-690,330, on phosphatidylinositol cycle markers". J. Pharmacol. Exp. Ther. 270 (1): 70–6. PMID 8035344.
- ^ King JS, Teo R, Ryves J, Reddy JV, Peters O, Orabi B, Hoeller O, Williams RS, Harwood AJ (2009). "The mood stabiliser lithium suppresses PIP3 signalling in Dictyostelium and human cells". Dis Models Mech. 2 (5–6): 306–12. doi:10.1242/dmm.001271. PMC 2675811. PMID 19383941.
- ^ Sarkar S, Rubinsztein DC (2006). "Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations". Autophagy. 2 (2): 132–4. doi:10.4161/auto.2387. PMID 16874097.
- ^ Williams RS, Cheng L, Mudge AW, Harwood AJ (May 2002). "A common mechanism of action for three mood-stabilizing drugs". Nature. 417 (6886): 292–5. Bibcode:2002Natur.417..292W. doi:10.1038/417292a. PMID 12015604. S2CID 4302048.
Further reading
- Bone R, Springer JP, Atack JR (1992). "Structure of inositol monophosphatase, the putative target of lithium therapy". Proc. Natl. Acad. Sci. U.S.A. 89 (21): 10031–10035. Bibcode:1992PNAS...8910031B. doi:10.1073/pnas.89.21.10031. PMC 50271. PMID 1332026.
- Hallcher LM, Sherman WR (1981). "The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain". J. Biol. Chem. 255 (22): 10896–901. doi:10.1016/S0021-9258(19)70391-3. PMID 6253491.
- Bone R, Frank L, Springer JP, Pollack SJ, Osborne S, Atack JR, Knowles MR, McAllister G, Ragan CI (1994). "Structural analysis of inositol monophosphatase complexes with substrates". Biochemistry. 33 (32): 9460–9467. doi:10.1021/bi00198a011. PMID 8068620.
- Bone R, Frank L, Springer JP, Atack JR (1994). "Structural studies of metal binding by inositol monophosphatase: evidence for two-metal ion catalysis". Biochemistry. 33 (32): 9468–9476. doi:10.1021/bi00198a012. PMID 8068621.
- Ganzhorn AJ, Lepage P, Pelton PD, Strasser F, Vincendon P, Rondeau JM (1996). "The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase". Biochemistry. 35 (33): 10957–10966. doi:10.1021/bi9603837. PMID 8718889.
- Parthasarathy L, Parthasarathy R, Vadnal R (1997). "Molecular characterization of coding and untranslated regions of rat cortex lithium-sensitive myo-inositol monophosphatase cDNA". Gene. 191 (1): 81–87. doi:10.1016/S0378-1119(97)00045-0. PMID 9210592.
- Sjøholt G, Molven A, Løvlie R, Wilcox A, Sikela JM, Steen VM (1997). "Genomic structure and chromosomal localization of a human myo-inositol monophosphatase gene (IMPA)". Genomics. 45 (1): 113–122. doi:10.1006/geno.1997.4862. PMID 9339367.
- Nemanov L, Ebstein RP, Belmaker RH, Osher Y, Agam G (1999). "Effect of bipolar disorder on lymphocyte inositol monophosphatase mRNA levels". The International Journal of Neuropsychopharmacology. 2 (1): 25–29. doi:10.1017/S1461145799001315. PMID 11281967. S2CID 16964945.
- Bahn JH, Kim AY, Jang SH, Lee BR, Ahn JY, Joo HM, Kan TC, Won MH, Kwon HY (2002). "Production of monoclonal antibodies and immunohistochemical studies of brain myo-inositol monophosphate phosphatase". Mol. Cells. 13 (1): 21–7. doi:10.1016/S1016-8478(23)14999-5. PMID 11911470.
- Atack JR, Schapiro MB (2002). "Inositol monophosphatase activity in normal, Down syndrome and dementia of the Alzheimer type CSF". Neurobiol. Aging. 23 (3): 389–396. doi:10.1016/S0197-4580(01)00335-9. PMID 11959401. S2CID 24701473.
- Berggard T, Szczepankiewicz O, Thulin E, Linse S (2003). "Myo-inositol monophosphatase is an activated target of calbindin D28k". J. Biol. Chem. 277 (44): 41954–41959. doi:10.1074/jbc.M203492200. PMID 12176979.
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–16903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Sjøholt G, Ebstein RP, Lie RT, Berle JØ, Mallet J, Deleuze JF, Levinson DF, Laurent C, Mujahed M (2005). "Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder". Mol. Psychiatry. 9 (6): 621–629. doi:10.1038/sj.mp.4001460. PMID 14699425. S2CID 28747842.
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–2127. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–1178. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Ohnishi T, Ohba H, Seo KC, Im J, Sato Y, Iwayama Y, Furuichi T, Chung SK, Yoshikawa T (2007). "Spatial expression patterns and biochemical properties distinguish a second myo-inositol monophosphatase IMPA2 from IMPA1". J. Biol. Chem. 282 (1): 637–646. doi:10.1074/jbc.M604474200. PMID 17068342.
External links
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Inositol monophosphatase 1
>Text je dostupný pod licencí Creative Commons Uveďte autora – Zachovejte licenci, případně za dalších podmínek. Podrobnosti naleznete na stránce Podmínky užití.
Protein Data Bank
Gene nomenclature
Mendelian Inheritance in Man
Mouse Genome Informatics
HomoloGene
GeneCards
Orthologous MAtrix
Enzyme Commission number
Human genome
File:Ideogram human chromosome 8.svg
Chromosome
Chromosome 8 (human)
File:Human chromosome 8 ideogram.svg
File:HSR 1996 II 3.5e.svg
File:Red rectangle 2x18.png
Locus (genetics)
Base pair
Base pair
Laboratory mouse
File:Ideogram house mouse chromosome 3.svg
Chromosome
File:Ideogram of house mouse chromosome 3.svg
File:HSR 1996 II 3.5e.svg
File:Red rectangle 2x18.png
Locus (genetics)
Base pair
Base pair
Gene expression
Human genome
Laboratory mouse
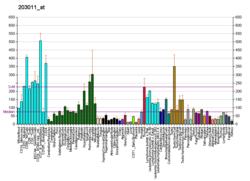
File:PBB GE IMPA1 203011 at fs.png
Gene ontology
Orthologs
Entrez
Ensembl
UniProt
PubMed
Wikidata
Q18027902
Q18258229
Enzyme
Gene
S100B
Calbindin
In vitro
Bioavailability
In vivo
Inositol
Pleiotropic
Phosphoinositide
Autophagy
Apoptosis
Lithium carbonate
Carbamazepine
Valproic acid
Phosphatidylinositol
In vitro
Ensembl genome database project
Ensembl genome database project
Doi (identifier)
PMC (identifier)
PMID (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Bibcode (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
PMID (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Bibcode (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
Bibcode (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
Doi (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
Doi (identifier)
PMID (identifier)
Bibcode (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
Doi (identifier)
PMC (identifier)
PMID (identifier)
Bibcode (identifier)
Doi (identifier)
PMID (identifier)
S2CID (identifier)
Doi (identifier)
PMID (identifier)
Template:PDB Gallery
Template talk:PDB Gallery
Special:EditPage/Template:PDB Gallery
File:PDB 1awb EBI.jpg
File:PDB 1ima EBI.jpg
File:PDB 1imb EBI.jpg
File:PDB 1imc EBI.jpg
File:PDB 1imd EBI.jpg
File:PDB 1ime EBI.jpg
File:PDB 1imf EBI.jpg
File:PDB 2hhm EBI.jpg
Template:Esterases
Template talk:Esterases
Special:EditPage/Template:Esterases
Hydrolase
Esterase
Enzyme Commission number
List of EC numbers (EC 3)#EC 3.1.1: Carboxylic Ester Hydrolases
Carboxylic acid
Cholinesterase
Acetylcholinesterase
Butyrylcholinesterase
Pectinesterase
6-phosphogluconolactonase
PAF acetylhydrolase
Lipase
Bile salt-dependent lipase
Gastric lipase
Lingual lipase
Pancreatic lipase
Lysosomal lipase
Hormone-sensitive lipase
Endothelial lipase
Hepatic lipase
Lipoprotein lipase
Monoacylglycerol lipase
Diacylglycerol lipase
Phospholipase
Phospholipase A1
Phospholipase A2
Phospholipase B
Cutinase
PETase
List of EC numbers (EC 3)#EC 3.1.2: Thioester Hydrolases
Thioesterase
Palmitoyl protein thioesterase
Ubiquitin carboxy-terminal hydrolase L1
4-hydroxybenzoyl-CoA thioesterase
List of EC numbers (EC 3)#EC 3.1.3: Phosphoric Monoester Hydrolases
Phosphatase
Alkaline phosphatase
ALPI
ALPL
Placental alkaline phosphatase
Acid phosphatase
Prostatic acid phosphatase
Tartrate-resistant acid phosphatase
Purple acid phosphatases
Nucleotidase
Glucose 6-phosphatase
Fructose 1,6-bisphosphatase
Calcineurin
Protein phosphatase
PP2A
OCRL
Pyruvate dehydrogenase phosphatase
Phosphofructokinase 2
PTEN (gene)
Phytase
Beta-propeller phytase
Inositol-phosphate phosphatase
Inositol monophosphatase 2
Inositol monophosphatase 3
Protein phosphatase
Protein tyrosine phosphatase
Protein serine/threonine phosphatase
Dual-specificity phosphatase
List of EC numbers (EC 3)#EC 3.1.4: Phosphoric Diester Hydrolases
Phosphodiesterase
Autotaxin
Phospholipase
Phospholipase C
Phospholipase D
Sphingomyelin phosphodiesterase
Sphingomyelin phosphodiesterase 1
PDE1
Phosphodiesterase 2
Phosphodiesterase 3
PDE4A
PDE4B
CGMP-specific phosphodiesterase type 5
Lecithinase
Clostridium perfringens alpha toxin
Cyclic nucleotide phosphodiesterase
List of EC numbers (EC 3)#EC 3.1.6: Sulfuric Ester Hydrolases
Sulfatase
Arylsulfatase
Arylsulfatase A
Arylsulfatase B
Arylsulfatase L
Steroid sulfatase
Galactosamine-6 sulfatase
Iduronate-2-sulfatase
N-acetylglucosamine-6-sulfatase
Nuclease
Deoxyribonuclease
Ribonuclease
List of EC numbers (EC 3)#EC 3.1.11: Exodeoxyribonucleases Producing 5.27-Phosphomonoesters
Exonuclease
Exodeoxyribonuclease
RecBCD
Exoribonuclease
Oligonucleotidase
List of EC numbers (EC 3)#EC 3.1.21: Endodeoxyribonucleases Producing 5.27-Phosphomonoesters
Endonuclease
Endodeoxyribonuclease
Deoxyribonuclease I
Deoxyribonuclease II
Deoxyribonuclease IV
Restriction enzyme
Template:Restriction enzyme
UvrABC endonuclease
Endoribonuclease
RNase III
Drosha
Microprocessor complex
Dicer
RNA-induced silencing complex
RNase H
RNASEH1
RNASEH2A
RNASEH2B
RNASEH2C
RNase P
Ribonuclease A
Ribonuclease Z
Ribonuclease E
RNASE1
Eosinophil-derived neurotoxin
Eosinophil cationic protein
Angiogenin
Ribonuclease T1
Nuclease S1
Mung bean nuclease
Serratia marcescens nuclease
Micrococcal nuclease
Template:Enzymes
Template talk:Enzymes
Special:EditPage/Template:Enzymes
Enzyme
Active site
Binding site
Catalytic triad
Oxyanion hole
Enzyme promiscuity
Diffusion-limited enzyme
Cofactor (biochemistry)
Enzyme catalysis
Allosteric regulation
Cooperativity
Enzyme inhibitor
Enzyme activator
Enzyme Commission number
Protein superfamily
Protein family
List of enzymes
Enzyme kinetics
Eadie–Hofstee diagram
Hanes–Woolf plot
Lineweaver–Burk plot
Michaelis–Menten kinetics
Oxidoreductase
List of EC numbers (EC 1)
Transferase
List of EC numbers (EC 2)
Hydrolase
List of EC numbers (EC 3)
Lyase
List of EC numbers (EC 4)
Isomerase
List of EC numbers (EC 5)
Ligase
List of EC numbers (EC 6)
Translocase
List of EC numbers (EC 7)
Wikipedia:Contents/Portals
File:Issoria lathonia.jpg
Portal:Biology
Inositol monophosphatase 1
Inositol monophosphatase 1
Main Page
Wikipedia:Contents
Updating...x
Text je dostupný za podmienok Creative
Commons Attribution/Share-Alike License 3.0 Unported; prípadne za ďalších
podmienok.
Podrobnejšie informácie nájdete na stránke Podmienky
použitia.