A | B | C | D | E | F | G | H | CH | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
| Human Y chromosome | |
|---|---|
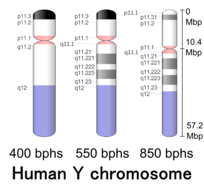
Human Y chromosome (after G-banding) | |
 Y chromosome in human male karyogram | |
| Features | |
| Length (bp) | 62,460,029 bp (CHM13) |
| No. of genes | 63 (CCDS)[1] |
| Type | Allosome |
| Centromere position | Acrocentric[2] (10.4 Mbp[3]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome Y |
| Entrez | Chromosome Y |
| NCBI | Chromosome Y |
| UCSC | Chromosome Y |
| Full DNA sequences | |
| RefSeq | NC_000024 (FASTA) |
| GenBank | CM000686 (FASTA) |
The Y chromosome is one of two sex chromosomes (allosomes) in therian mammals, including humans, and many other animals. The other is the X chromosome. Y is normally the sex-determining chromosome in many species, since it is the presence or absence of Y that determines the male or female sex of offspring produced in sexual reproduction. In mammals, the Y chromosome contains the gene SRY, which triggers male development. The DNA in the human Y chromosome is composed of about 59 million base pairs, making it similar in size to chromosome 19.[4] The Y chromosome is passed only from father to son. With a 30% difference between humans and chimpanzees, the Y chromosome is one of the fastest-evolving parts of the human genome.[5] The human Y chromosome carries 693 genes, with 107 of these being protein-coding, but some genes are repeated and that makes the number of exclusive protein-coding genes just 42, the numbers are given for telomere-to-telomere CHM13.[6] CCDS only classified 63 out of 107. All single-copy Y-linked genes are hemizygous (present on only one chromosome) except in cases of aneuploidy such as XYY syndrome or XXYY syndrome.
Overview
Discovery
The Y chromosome was identified as a sex-determining chromosome by Nettie Stevens at Bryn Mawr College in 1905 during a study of the mealworm Tenebrio molitor. Edmund Beecher Wilson independently discovered the same mechanisms the same year, working with hemiptera. Stevens proposed that chromosomes always existed in pairs and that the smaller chromosome (now labelled "Y") was the pair of the X chromosome discovered in 1890 by Hermann Henking. She realized that the previous idea of Clarence Erwin McClung, that the X chromosome determines sex, was wrong and that sex determination is, in fact, due to the presence or absence of the Y chromosome. In the early 1920s Theophilus Painter determined that X and Y chromosomes determined sex in humans (and other mammals).[7]
The chromosome was given the name "Y" simply to follow on from Henking's "X" alphabetically.[8][9] The idea that the Y chromosome was named after its similarity in appearance to the letter "Y" is mistaken. All chromosomes normally appear as an amorphous blob under the microscope and only take on a well-defined shape during mitosis. This shape is vaguely X-shaped for all chromosomes. It is entirely coincidental that the Y chromosome, during mitosis, has two very short branches which can look merged under the microscope and appear as the descender of a Y-shape.[10]
Variations
Most therian mammals have only one pair of sex chromosomes in each cell. Males have one Y chromosome and one X chromosome, while females have two X chromosomes. In mammals, the Y chromosome contains a gene, SRY, which triggers embryonic development as a male. The Y chromosomes of humans and other mammals also contain other genes needed for normal sperm production.
There are exceptions, however. Among humans, some men have two Xs and a Y ("XXY", see Klinefelter syndrome), or one X and two Ys (see XYY syndrome), and some women have three Xs or a single X instead of a double X ("X0", see Turner syndrome). There are other exceptions in which SRY is damaged (leading to an XY female), or copied to the X (leading to an XX male).
Origins and evolution
Before Y chromosome
Many ectothermic vertebrates have no sex chromosomes. If they have different sexes, sex is determined environmentally rather than genetically. For some of them, especially reptiles, sex depends on the incubation temperature. Some vertebrates are hermaphrodites, although other than a very few ray-finned fish, they are sequential (the same organism produces male or female gametes, but never both, at different points in its life), rather than simultaneous (the same organism producing both male and female gametes at the same time).
Origin
The X and Y chromosomes are thought to have evolved from a pair of identical chromosomes,[11][12] termed autosomes, when an ancestral animal developed an allelic variation, a so-called "sex locus" – simply possessing this allele caused the organism to be male.[13] The chromosome with this allele became the Y chromosome, while the other member of the pair became the X chromosome. Over time, genes that were beneficial for males and harmful to (or had no effect on) females either developed on the Y chromosome or were acquired through the process of translocation.[14]
Until recently, the X and Y chromosomes were thought to have diverged around 300 million years ago.[15] However, research published in 2010,[16] and particularly research published in 2008 documenting the sequencing of the platypus genome,[17] has suggested that the XY sex-determination system would not have been present more than 166 million years ago, at the split of the monotremes from other mammals.[18] This re-estimation of the age of the therian XY system is based on the finding that sequences that are on the X chromosomes of marsupials and eutherian mammals are present on the autosomes of platypus and birds.[18] The older estimate was based on erroneous reports that the platypus X chromosomes contained these sequences.[19][20]
Recombination inhibition
Recombination between the X and Y chromosomes proved harmful—it resulted in males without necessary genes formerly found on the Y chromosome, and females with unnecessary or even harmful genes previously only found on the Y chromosome. As a result, genes beneficial to males accumulated near the sex-determining genes, and recombination in this region was suppressed in order to preserve this male specific region.[13] Over time, the Y chromosome changed in such a way as to inhibit the areas around the sex determining genes from recombining at all with the X chromosome. As a result of this process, 95% of the human Y chromosome is unable to recombine. Only the tips of the Y and X chromosomes recombine. The tips of the Y chromosome that could recombine with the X chromosome are referred to as the pseudoautosomal region. The rest of the Y chromosome is passed on to the next generation intact, allowing for its use in tracking human evolution.[citation needed]
Degeneration
By one estimate, the human Y chromosome has lost 1,393 of its 1,438 original genes over the course of its existence, and linear extrapolation of this 1,393-gene loss over 300 million years gives a rate of genetic loss of 4.6 genes per million years.[21] Continued loss of genes at the rate of 4.6 genes per million years would result in a Y chromosome with no functional genes – that is the Y chromosome would lose complete function – within the next 10 million years, or half that time with the current age estimate of 160 million years.[13][22] Comparative genomic analysis reveals that many mammalian species are experiencing a similar loss of function in their heterozygous sex chromosome. Degeneration may simply be the fate of all non-recombining sex chromosomes, due to three common evolutionary forces: high mutation rate, inefficient selection, and genetic drift.[13]
However, comparisons of the human and chimpanzee Y chromosomes (first published in 2005) show that the human Y chromosome has not lost any genes since the divergence of humans and chimpanzees between 6–7 million years ago,[23] and a scientific report in 2012 stated that only one gene had been lost since humans diverged from the rhesus macaque 25 million years ago.[24] These facts provide direct evidence that the linear extrapolation model is flawed and suggest that the current human Y chromosome is either no longer shrinking or is shrinking at a much slower rate than the 4.6 genes per million years estimated by the linear extrapolation model.
High mutation rate
The human Y chromosome is particularly exposed to high mutation rates due to the environment in which it is housed. The Y chromosome is passed exclusively through sperm, which undergo multiple cell divisions during gametogenesis. Each cellular division provides further opportunity to accumulate base pair mutations. Additionally, sperm are stored in the highly oxidative environment of the testis, which encourages further mutation. These two conditions combined put the Y chromosome at a greater opportunity of mutation than the rest of the genome.[13] The increased mutation opportunity for the Y chromosome is reported by Graves as a factor 4.8.[13] However, her original reference obtains this number for the relative mutation rates in male and female germ lines for the lineage leading to humans.[25]
The observation that the Y chromosome experiences little meiotic recombination and has an accelerated rate of mutation and degradative change compared to the rest of the genome suggests an evolutionary explanation for the adaptive function of meiosis with respect to the main body of genetic information. Brandeis[26] proposed that the basic function of meiosis (particularly meiotic recombination) is the conservation of the integrity of the genome, a proposal consistent with the idea that meiosis is an adaptation for repairing DNA damage.[27]
Inefficient selection
Without the ability to recombine during meiosis, the Y chromosome is unable to expose individual alleles to natural selection. Deleterious alleles are allowed to "hitchhike" with beneficial neighbors, thus propagating maladapted alleles into the next generation. Conversely, advantageous alleles may be selected against if they are surrounded by harmful alleles (background selection). Due to this inability to sort through its gene content, the Y chromosome is particularly prone to the accumulation of "junk" DNA. Massive accumulations of retrotransposable elements are scattered throughout the Y.[13] The random insertion of DNA segments often disrupts encoded gene sequences and renders them nonfunctional. However, the Y chromosome has no way of weeding out these "jumping genes". Without the ability to isolate alleles, selection cannot effectively act upon them.[citation needed]
A clear, quantitative indication of this inefficiency is the entropy rate of the Y chromosome. Whereas all other chromosomes in the human genome have entropy rates of 1.5–1.9 bits per nucleotide (compared to the theoretical maximum of exactly 2 for no redundancy), the Y chromosome's entropy rate is only 0.84.[28] This means the Y chromosome has a much lower information content relative to its overall length; it is more redundant.
Genetic drift
Even if a well adapted Y chromosome manages to maintain genetic activity by avoiding mutation accumulation, there is no guarantee it will be passed down to the next generation. The population size of the Y chromosome is inherently limited to 1/4 that of autosomes: diploid organisms contain two copies of autosomal chromosomes while only half the population contains 1 Y chromosome. Thus, genetic drift is an exceptionally strong force acting upon the Y chromosome. Through sheer random assortment, an adult male may never pass on his Y chromosome if he only has female offspring. Thus, although a male may have a well adapted Y chromosome free of excessive mutation, it may never make it into the next gene pool.[13] The repeat random loss of well-adapted Y chromosomes, coupled with the tendency of the Y chromosome to evolve to have more deleterious mutations rather than less for reasons described above, contributes to the species-wide degeneration of Y chromosomes through Muller's ratchet.[29]
Gene conversion
As it has been already mentioned, the Y chromosome is unable to recombine during meiosis like the other human chromosomes; however, in 2003, researchers from MIT discovered a process which may slow down the process of degradation. They found that human Y chromosome is able to "recombine" with itself, using palindrome base pair sequences.[30] Such a "recombination" is called gene conversion.
In the case of the Y chromosomes, the palindromes are not noncoding DNA; these strings of bases contain functioning genes important for male fertility. Most of the sequence pairs are greater than 99.97% identical. The extensive use of gene conversion may play a role in the ability of the Y chromosome to edit out genetic mistakes and maintain the integrity of the relatively few genes it carries. In other words, since the Y chromosome is single, it has duplicates of its genes on itself instead of having a second, homologous, chromosome. When errors occur, it can use other parts of itself as a template to correct them.[31]
Findings were confirmed by comparing similar regions of the Y chromosome in humans to the Y chromosomes of chimpanzees, bonobos and gorillas. The comparison demonstrated that the same phenomenon of gene conversion appeared to be at work more than 5 million years ago, when humans and the non-human primates diverged from each other.[31]
Future evolution
According to some theories, in the terminal stages of the degeneration of the Y chromosome, other chromosomes increasingly take over genes and functions formerly associated with it and finally, within the framework of this theory, finally, the Y chromosome disappears entirely, and a new sex-determining system arises.[13][neutrality is disputed][improper synthesis?] Several species of rodent in the sister families Muridae and Cricetidae have reached these stages,[32][33] in the following ways:
- The Transcaucasian mole vole, Ellobius lutescens, the Zaisan mole vole, Ellobius tancrei, and the Japanese spinous country rats Tokudaia osimensis and Tokudaia tokunoshimensis, have lost the Y chromosome and SRY entirely.[13][34][35] Tokudaia spp. have relocated some other genes ancestrally present on the Y chromosome to the X chromosome.[35] Both sexes of Tokudaia spp. and Ellobius lutescens have an XO genotype (Turner syndrome),[35] whereas all Ellobius tancrei possess an XX genotype.[13] The new sex-determining system(s) for these rodents remains unclear.
- The wood lemming Myopus schisticolor, the Arctic lemming, Dicrostonyx torquatus, and multiple species in the grass mouse genus Akodon have evolved fertile females who possess the genotype generally coding for males, XY, in addition to the ancestral XX female, through a variety of modifications to the X and Y chromosomes.[32][36][37]
- In the creeping vole, Microtus oregoni, the females, with just one X chromosome each, produce X gametes only, and the males, XY, produce Y gametes, or gametes devoid of any sex chromosome, through nondisjunction.[38]
Outside of the rodents, the black muntjac, Muntiacus crinifrons, evolved new X and Y chromosomes through fusions of the ancestral sex chromosomes and autosomes.[39]
Modern data cast doubt on this hypothesis.[40] This conclusion was reached by scientists who studied the Y chromosomes of rhesus monkeys. When genomically comparing the Y chromosome of rhesus monkeys and humans, scientists found very few differences, given that humans and rhesus monkeys diverged 30 million years ago.[41]
Some organisms have lost the Y chromosome. For example, most species of Nematodes. However, in order for the complete elimination of Y to occur, it was necessary to develop an alternative way of determining sex (for example, by determining sex by the ratio of the X chromosome to autosomes), and any genes necessary for male function had to be moved to other chromosomes.[40] In the meantime, modern data demonstrate the complex mechanisms of Y chromosome evolution and the fact that the disappearance of the Y chromosome is not guaranteed.
1:1 sex ratio
Fisher's principle outlines why almost all species using sexual reproduction have a sex ratio of 1:1. W. D. Hamilton gave the following basic explanation in his 1967 paper on "Extraordinary sex ratios",[42] given the condition that males and females cost equal amounts to produce:
- Suppose male births are less common than female.
- A newborn male then has better mating prospects than a newborn female, and therefore can expect to have more offspring.
- Therefore, parents genetically disposed to produce males tend to have more than average numbers of grandchildren born to them.
- Therefore, the genes for male-producing tendencies spread, and male births become more common.
- As the 1:1 sex ratio is approached, the advantage associated with producing males dies away.
- The same reasoning holds if females are substituted for males throughout. Therefore, 1:1 is the equilibrium ratio.
Non-therian Y chromosome
Many groups of organisms in addition to therian mammals have Y chromosomes, but these Y chromosomes do not share common ancestry with therian Y chromosomes. Such groups include monotremes, Drosophila, some other insects, some fish, some reptiles, and some plants. In Drosophila melanogaster, the Y chromosome does not trigger male development. Instead, sex is determined by the number of X chromosomes. The D. melanogaster Y chromosome does contain genes necessary for male fertility. So XXY D. melanogaster are female, and D. melanogaster with a single X (X0), are male but sterile. There are some species of Drosophila in which X0 males are both viable and fertile.[citation needed]
ZW chromosomes
Other organisms have mirror image sex chromosomes: where the homogeneous sex is the male, said to have two Z chromosomes, and the female is the heterogeneous sex with a Z chromosome and a W chromosome.[43] For example, female birds, snakes, and butterflies have ZW sex chromosomes, and males have ZZ sex chromosomes.[43][44][45]
Non-inverted Y chromosome
There are some species, such as the Japanese rice fish, in which the XY system is still developing and cross over between the X and Y is still possible. Because the male specific region is very small and contains no essential genes, it is even possible to artificially induce XX males and YY females to no ill effect.[46]
Multiple XY pairs
Monotremes possess four or five (platypus) pairs of XY sex chromosomes, each pair consisting of sex chromosomes with homologous regions. The chromosomes of neighboring pairs are partially homologous, such that a chain is formed during mitosis.[19] The first X chromosome in the chain is also partially homologous with the last Y chromosome, indicating that profound rearrangements, some adding new pieces from autosomes, have occurred in history.[47][48]: fig. 5
Platypus sex chromosomes have strong sequence similarity with the avian Z chromosome, (indicating close homology),[17] and the SRY gene so central to sex-determination in most other mammals is apparently not involved in platypus sex-determination.[18]
Human Y chromosome
This section may require cleanup to meet Wikipedia's quality standards. The specific problem is: Too many subsections. Article might benefit from moving h3 subsections into h2 sections, if we can somehow reconcile the gap between all therians and humans. "Origins and evolution" section has a human focus, but the discussion does include all therians. (October 2021) |
In humans, the Y chromosome spans about 58 million base pairs (the building blocks of DNA) and represents almost 2% of the total DNA in a male cell.[49] The human Y chromosome contains over 200 genes, at least 72 of which code for proteins.[4] Traits that are inherited via the Y chromosome are called Y-linked traits, or holandric traits (from Ancient Greek ὅλος hólos, "whole" + ἀνδρός andrós, "male").[50]
Loss of Y chromosome
Men can lose the Y chromosome in a subset of cells, which is called the mosaic loss of chromosome Y (mLOY). This post-zygotic mutation is strongly associated with age, affecting about 15%[contradictory] of men 70 years of age. Smoking is another important risk factor for LOY.[51] It has been found that men with a higher percentage of hematopoietic stem cells in blood lacking the Y chromosome (and perhaps a higher percentage of other cells lacking it) have a higher risk of certain cancers and have a shorter life expectancy. Men with LOY (which was defined as no Y in at least 18% of their hematopoietic cells) have been found to die 5.5 years earlier on average than others. This has been interpreted as a sign that the Y chromosome plays a role going beyond sex determination and reproduction.[52] However, it was thought that the loss of Y could also be an effect rather than a cause and/or a "neutral karyotype related to normal aging".[53] In 2022, a study showed that blood cells' loss of the Y chromosome in a subset of cells (mLOY), reportedly affecting at least 40%[contradictory] of 70 years-old men to some degree, contributes to fibrosis, heart risks, and mortality in a causal way.[54] Male smokers have between 1.5 and 2 times the risk of non-respiratory cancers as female smokers.[55][56] Potential countermeasures identified so far include not smoking or stopping smoking and at least one potential drug that "may help counteract the harmful effects of the chromosome loss" is under investigation.[57][58][better source needed]
Structure
This article is missing information about NRY/MSY structure - How there's a huge chunk of heterochromatin in q, nomenclature of the palindromes and amplicons, TTTY transcripts, etc. Best if we add a figure that mashes together the tops of Colaco 2018 Fig 1 and PMID 12815422 fig 3.. (October 2021) |
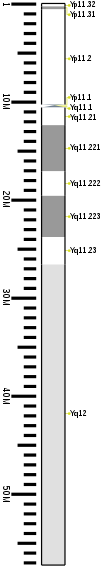
Cytogenetic band
| Chr. | Arm[63] | Band[64] | ISCN start[65] |
ISCN stop[65] |
Basepair start |
Basepair stop |
Stain[66] | Density |
|---|---|---|---|---|---|---|---|---|
| Y | p | 11.32 | 0 | 149 | 1 | 300,000 | gneg | |
| Y | p | 11.31 | 149 | 298 | 300,001 | 600,000 | gpos | 50 |
| Y | p | 11.2 | 298 | 1043 | 600,001 | 10,300,000 | gneg | |
| Y | p | 11.1 | 1043 | 1117 | 10,300,001 | 10,400,000 | acen | |
| Y | q | 11.1 | 1117 | 1266 | 10,400,001 | 10,600,000 | acen | |
| Y | q | 11.21 | 1266 | 1397 | 10,600,001 | 12,400,000 | gneg | |
| Y | q | 11.221 | 1397 | 1713 | 12,400,001 | 17,100,000 | gpos | 50 |
| Y | q | 11.222 | 1713 | 1881 | 17,100,001 | 19,600,000 | gneg | |
| Y | q | 11.223 | 1881 | 2160 | 19,600,001 | 23,800,000 | gpos | 50 |
| Y | q | 11.23 | 2160 | 2346 | 23,800,001 | 26,600,000 | gneg | |
| Y | q | 12 | 2346 | 3650 | 26,600,001 | 57,227,415 | gvar |
Non-combining region of Y (NRY)
The human Y chromosome is normally unable to recombine with the X chromosome, except for small pieces of pseudoautosomal regions (PARs) at the telomeres (which comprise about 5% of the chromosome's length). These regions are relics of ancient homology between the X and Y chromosomes. The bulk of the Y chromosome, which does not recombine, is called the "NRY", or non-recombining region of the Y chromosome.[67] Single-nucleotide polymorphisms (SNPs) in this region are used to trace direct paternal ancestral lines.
More specifically, PAR1 is at 0.1–2.7 Mb. PAR2 is at 56.9–57.2 Mb. The non-recombining region (NRY) or male-specific region (MSY) sits between.
Sequence classes
Genesedit
Number of genesedit
The following are some of the gene count estimates of human Y chromosome. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[68]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 63 | — | — | [1] | 2016-09-08 |
| HGNC | 45 | 55 | 381 | [69] | 2017-05-12 |
| Ensembl | 63 | 109 | 392 | [70] | 2017-03-29 |
| UniProt | 47 | — | — | [71] | 2018-02-28 |
| NCBI | 73 | 122 | 400 | [72][73][74] | 2017-05-19 |
Gene listedit
In general, the human Y chromosome is extremely gene poor—it is one of the largest gene deserts in the human genome. Disregarding pseudoautosomal genes, genes encoded on the human Y chromosome include:
| Name | X paralog | Note |
|---|---|---|
| SRY | SOX3 | Sex-determining region. This is the short p arm Yp. |
| ZFY | ZFX | Zinc finger. |
| RPS4Y1 | RPS4X | Ribosomal protein S4. |
| AMELY | AMELX | Amelogenin. |
| TBL1Y | TBL1X | |
| PCDH11Y | PDCH11X | X-transposed region (XTR) from Xq21, one of two genes. Once dubbed "PAR3"[76] but later refuted.[77] |
| TGIF2LY | TGIF2LX | The other X-transposed gene. |
| TSPY1, TSPY2 | TSPX | Testis-specific protein. |
| AZFa | (none) | Not a gene. First part of the AZF region on arm q. Contains the four following genes. X counterparts escape inactivation. |
| USP9Y | USP9X | Ubiquitin protease. |
| DDX3Y | DDX3X | Helicase. |
| UTY | UTX | Histone demethylase. |
| TB4Y | TB4X | |
| AZFb | (none) | Second AZF region on arm q. Prone to NAHR non-allelic homologous recombination with AZFc. Overlaps with AZFc. Contains three single-copy gene regions and repeats. |
| CYorf15 | CXorf15 | |
| RPS4Y2 | RPS4X | Another copy of ribosomal protein S4. |
| EIF1AY | EIF4AX | |
| KDM5D | KDM5C | |
| XKRY | XK (protein) | Found in the "yellow" amplicon. |
| HSFY1, HSFY2 | HSFX1, HSFX2 | Found in the "blue" amplicon. |
| PRY, PRY2 | Found in the "blue" amplicon. Identified by similarity to PTPN13 (Chr. 4). | |
| RBMY1A1 | RBMY | Large number of copies. Part of an RBM gene family of RNA recognition motif (RRM) proteins. |
| AZFc | (none) | Final (distal) part of the AZF. Multiple palindromes. |
| DAZ1, DAZ2, DAZ3, DAZ4 | RRM genes in two palindromic clusters. BOLL and DAZLA are autosomal homologs. | |
| CDY1, CDY2 | CDY1 is actually two identical copies. CDY2 is two closely related copies in palindrome P5. Probably derived from autosomal CDYL. | |
| VCY1, VCY2 | VCX1 through 3 | Three copies of VCX2 (BPY2). Part of the VCX/VCY family. The two copies of BPY1 are instead in Yq11.221/AZFa. |
Y-chromosome-linked diseasesedit
Diseases linked to the Y chromosome typically involve an aneuploidy, an atypical number of chromosomes.
Y chromosome microdeletionedit
Y chromosome microdeletion (YCM) is a family of genetic disorders caused by missing genes in the Y chromosome. Many affected men exhibit no symptoms and lead normal lives. However, YCM is also known to be present in a significant number of men with reduced fertility or reduced sperm count.[citation needed]
Defective Y chromosomeedit
This results in the person presenting a female phenotype (i.e., is born with female-like genitalia) even though that person possesses an XY karyotype. The lack of the second X results in infertility. In other words, viewed from the opposite direction, the person goes through defeminization but fails to complete masculinization.[citation needed]
Zdroj:https://en.wikipedia.org?pojem=Y-DNA
>Text je dostupný pod licencí Creative Commons Uveďte autora – Zachovejte licenci, případně za dalších podmínek. Podrobnosti naleznete na stránce Podmínky užití.
Text je dostupný za podmienok Creative
Commons Attribution/Share-Alike License 3.0 Unported; prípadne za ďalších
podmienok.
Podrobnejšie informácie nájdete na stránke Podmienky
použitia.