A | B | C | D | E | F | G | H | CH | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9

Amino acids are organic compounds that contain both amino and carboxylic acid functional groups.[1] Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins.[2] Only these 22 appear in the genetic code of life.[3][4]
Amino acids can be classified according to the locations of the core structural functional groups (alpha- (α-), beta- (β-), gamma- (γ-) amino acids, etc.), other categories relate to polarity, ionization, and side chain group type (aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino acid residues form the second-largest component (water being the largest) of human muscles and other tissues.[5] Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence.
Amino acids are formally named by the IUPAC-IUBMB Joint Commission on Biochemical Nomenclature in terms of the fictitious "neutral" structure shown in the illustration. For example, the systematic name of alanine is 2-aminopropanoic acid, based on the formula CH3−CH(NH2)−COOH. The Commission justified this approach as follows:[6]
The systematic names and formulas given refer to hypothetical forms in which amino groups are unprotonated and carboxyl groups are undissociated. This convention is useful to avoid various nomenclatural problems but should not be taken to imply that these structures represent an appreciable fraction of the amino-acid molecules.
History
The first few amino acids were discovered in the early 1800s.[7][8] In 1806, French chemists Louis-Nicolas Vauquelin and Pierre Jean Robiquet isolated a compound from asparagus that was subsequently named asparagine, the first amino acid to be discovered.[9][10] Cystine was discovered in 1810,[11] although its monomer, cysteine, remained undiscovered until 1884.[12][10][a] Glycine and leucine were discovered in 1820.[13] The last of the 20 common amino acids to be discovered was threonine in 1935 by William Cumming Rose, who also determined the essential amino acids and established the minimum daily requirements of all amino acids for optimal growth.[14][15]
The unity of the chemical category was recognized by Wurtz in 1865, but he gave no particular name to it.[16] The first use of the term "amino acid" in the English language dates from 1898,[17] while the German term, Aminosäure, was used earlier.[18] Proteins were found to yield amino acids after enzymatic digestion or acid hydrolysis. In 1902, Emil Fischer and Franz Hofmeister independently proposed that proteins are formed from many amino acids, whereby bonds are formed between the amino group of one amino acid with the carboxyl group of another, resulting in a linear structure that Fischer termed "peptide".[19]
General structure
2-, alpha-, or α-amino acids[20] have the generic formula H2NCHRCOOH in most cases,[b] where R is an organic substituent known as a "side chain".[21]
Of the many hundreds of described amino acids, 22 are proteinogenic ("protein-building").[22][23][24] It is these 22 compounds that combine to give a vast array of peptides and proteins assembled by ribosomes.[25] Non-proteinogenic or modified amino acids may arise from post-translational modification or during nonribosomal peptide synthesis.
Chirality
The carbon atom next to the carboxyl group is called the α–carbon. In proteinogenic amino acids, it bears the amine and the R group or side chain specific to each amino acid. With four distinct substituents, the α–carbon is stereogenic in all α-amino acids except glycine. All chiral proteogenic amino acids have the L configuration. They are "left-handed" enantiomers, which refers to the stereoisomers of the alpha carbon.
A few D-amino acids ("right-handed") have been found in nature, e.g., in bacterial envelopes, as a neuromodulator (D-serine), and in some antibiotics.[26][27] Rarely, D-amino acid residues are found in proteins, and are converted from the L-amino acid as a post-translational modification.[28][c]
Side chains
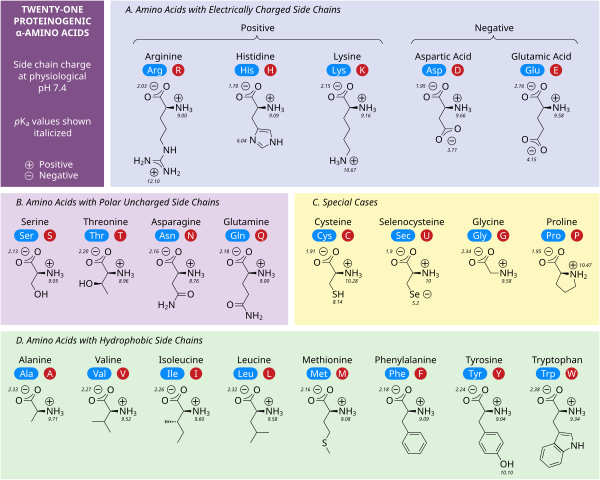
Polar charged side chains
Five amino acids possess a charge at neutral pH. Often these side chains appear at the surfaces on proteins to enable their solubility in water, and side chains with opposite charges form important electrostatic contacts called salt bridges that maintain structures within a single protein or between interfacing proteins.[31] Many proteins bind metal into their structures specifically, and these interactions are commonly mediated by charged side chains such as aspartate, glutamate and histidine. Under certain conditions, each ion-forming group can be charged, forming double salts.[32]
The two negatively charged amino acids at neutral pH are aspartate (Asp, D) and glutamate (Glu, E). The anionic carboxylate groups behave as Brønsted bases in most circumstances.[31] Enzymes in very low pH environments, like the aspartic protease pepsin in mammalian stomachs, may have catalytic aspartate or glutamate residues that act as Brønsted acids.

There are three amino acids with side chains that are cations at neutral pH: arginine (Arg, R), lysine (Lys, K) and histidine (His, H). Arginine has a charged guanidino group and lysine a charged alkyl amino group, and are fully protonated at pH 7. Histidine's imidazole group has a pKa of 6.0, and is only around 10 % protonated at neutral pH. Because histidine is easily found in its basic and conjugate acid forms it often participates in catalytic proton transfers in enzyme reactions.[31]
Polar uncharged side chains
The polar, uncharged amino acids serine (Ser, S), threonine (Thr, T), asparagine (Asn, N) and glutamine (Gln, Q) readily form hydrogen bonds with water and other amino acids.[31] They do not ionize in normal conditions, a prominent exception being the catalytic serine in serine proteases. This is an example of severe perturbation, and is not characteristic of serine residues in general. Threonine has two chiral centers, not only the L (2S) chiral center at the α-carbon shared by all amino acids apart from achiral glycine, but also (3R) at the β-carbon. The full stereochemical specification is (2S,3R)-L-threonine.
Hydrophobic side chains
Nonpolar amino acid interactions are the primary driving force behind the processes that fold proteins into their functional three dimensional structures.[31] None of these amino acids' side chains ionize easily, and therefore do not have pKas, with the exception of tyrosine (Tyr, Y). The hydroxyl of tyrosine can deprotonate at high pH forming the negatively charged phenolate. Because of this one could place tyrosine into the polar, uncharged amino acid category, but its very low solubility in water matches the characteristics of hydrophobic amino acids well.
Special case side chains
Several side chains are not described well by the charged, polar and hydrophobic categories. Glycine (Gly, G) could be considered a polar amino acid since its small size means that its solubility is largely determined by the amino and carboxylate groups. However, the lack of any side chain provides glycine with a unique flexibility among amino acids with large ramifications to protein folding.[31] Cysteine (Cys, C) can also form hydrogen bonds readily, which would place it in the polar amino acid category, though it can often be found in protein structures forming covalent bonds, called disulphide bonds, with other cysteines. These bonds influence the folding and stability of proteins, and are essential in the formation of antibodies. Proline (Pro, P) has an alkyl side chain and could be considered hydrophobic, but because the side chain joins back onto the alpha amino group it becomes particularly inflexible when incorporated into proteins. Similar to glycine this influences protein structure in a way unique among amino acids. Selenocysteine (Sec, U) is a rare amino acid not directly encoded by DNA, but is incorporated into proteins via the ribosome. Selenocysteine has a lower redox potential compared to the similar cysteine, and participates in several unique enzymatic reactions.[33] Pyrrolysine (Pyl, O) is another amino acid not encoded in DNA, but synthesized into protein by ribosomes.[34] It is found in archaeal species where it participates in the catalytic activity of several methyltransferases.
β- and γ-amino acids
Amino acids with the structure NH+3−CXY−CXY−CO−2, such as β-alanine, a component of carnosine and a few other peptides, are β-amino acids. Ones with the structure NH+3−CXY−CXY−CXY−CO−2 are γ-amino acids, and so on, where X and Y are two substituents (one of which is normally H).[6]
Zwitterions

The common natural forms of amino acids have a zwitterionic structure, with −NH+3 (−NH+2− in the case of proline) and −CO−2 functional groups attached to the same C atom, and are thus α-amino acids, and are the only ones found in proteins during translation in the ribosome. In aqueous solution at pH close to neutrality, amino acids exist as zwitterions, i.e. as dipolar ions with both NH+3 and CO−2 in charged states, so the overall structure is NH+3−CHR−CO−2. At physiological pH the so-called "neutral forms" −NH2−CHR−CO2H are not present to any measurable degree.[35] Although the two charges in the zwitterion structure add up to zero it is misleading to call a species with a net charge of zero "uncharged".
In strongly acidic conditions (pH below 3), the carboxylate group becomes protonated and the structure becomes an ammonio carboxylic acid, NH+3−CHR−CO2H. This is relevant for enzymes like pepsin that are active in acidic environments such as the mammalian stomach and lysosomes, but does not significantly apply to intracellular enzymes. In highly basic conditions (pH greater than 10, not normally seen in physiological conditions), the ammonio group is deprotonated to give NH2−CHR−CO−2.
Although various definitions of acids and bases are used in chemistry, the only one that is useful for chemistry in aqueous solution is that of Brønsted:[36][37] an acid is a species that can donate a proton to another species, and a base is one that can accept a proton. This criterion is used to label the groups in the above illustration. The carboxylate side chains of aspartate and glutamate residues are the principal Brønsted bases in proteins. Likewise, lysine, tyrosine and cysteine will typically act as a Brønsted acid. Histidine under these conditions can act both as a Brønsted acid and a base.
Isoelectric point

For amino acids with uncharged side-chains the zwitterion predominates at pH values between the two pKa values, but coexists in equilibrium with small amounts of net negative and net positive ions. At the midpoint between the two pKa values, the trace amount of net negative and trace of net positive ions balance, so that average net charge of all forms present is zero.[38] This pH is known as the isoelectric point pI, so pI = 1/2(pKa1 + pKa2).
For amino acids with charged side chains, the pKa of the side chain is involved. Thus for aspartate or glutamate with negative side chains, the terminal amino group is essentially entirely in the charged form −NH+3, but this positive charge needs to be balanced by the state with just one C-terminal carboxylate group is negatively charged. This occurs halfway between the two carboxylate pKa values: pI = 1/2(pKa1 + pKa(R)), where pKa(R) is the side chain pKa.[37]
Similar considerations apply to other amino acids with ionizable side-chains, including not only glutamate (similar to aspartate), but also cysteine, histidine, lysine, tyrosine and arginine with positive side chains.
Amino acids have zero mobility in electrophoresis at their isoelectric point, although this behaviour is more usually exploited for peptides and proteins than single amino acids. Zwitterions have minimum solubility at their isoelectric point, and some amino acids (in particular, with nonpolar side chains) can be isolated by precipitation from water by adjusting the pH to the required isoelectric point.
Physicochemical properties
The 20 canonical amino acids can be classified according to their properties. Important factors are charge, hydrophilicity or hydrophobicity, size, and functional groups.[27] These properties influence protein structure and protein–protein interactions. The water-soluble proteins tend to have their hydrophobic residues (Leu, Ile, Val, Phe, and Trp) buried in the middle of the protein, whereas hydrophilic side chains are exposed to the aqueous solvent. (In biochemistry, a residue refers to a specific monomer within the polymeric chain of a polysaccharide, protein or nucleic acid.) The integral membrane proteins tend to have outer rings of exposed hydrophobic amino acids that anchor them in the lipid bilayer. Some peripheral membrane proteins have a patch of hydrophobic amino acids on their surface that sticks to the membrane. In a similar fashion, proteins that have to bind to positively charged molecules have surfaces rich in negatively charged amino acids such as glutamate and aspartate, while proteins binding to negatively charged molecules have surfaces rich in positively charged amino acids like lysine and arginine. For example, lysine and arginine are present in large amounts in the low-complexity regions of nucleic-acid binding proteins.[39] There are various hydrophobicity scales of amino acid residues.[40]
Some amino acids have special properties. Cysteine can form covalent disulfide bonds to other cysteine residues. Proline forms a cycle to the polypeptide backbone, and glycine is more flexible than other amino acids.
Glycine and proline are strongly present within low complexity regions of both eukaryotic and prokaryotic proteins, whereas the opposite is the case with cysteine, phenylalanine, tryptophan, methionine, valine, leucine, isoleucine, which are highly reactive, or complex, or hydrophobic.[39][41][42]
Many proteins undergo a range of posttranslational modifications, whereby additional chemical groups are attached to the amino acid residue side chains sometimes producing lipoproteins (that are hydrophobic),[43] or glycoproteins (that are hydrophilic)[44] allowing the protein to attach temporarily to a membrane. For example, a signaling protein can attach and then detach from a cell membrane, because it contains cysteine residues that can have the fatty acid palmitic acid added to them and subsequently removed.[45]
Table of standard amino acid abbreviations and properties
Although one-letter symbols are included in the table, IUPAC–IUBMB recommend[6] that "Use of the one-letter symbols should be restricted to the comparison of long sequences".
The one-letter notation was chosen by IUPAC-IUB based on the following rules:[46]
- Initial letters are used where there is no ambuiguity: C cysteine, H histidine, I isoleucine, M methionine, S serine, V valine,[46]
- Where arbitrary assignment is needed, the structurally simpler amino acids are given precedence: A Alanine, G glycine, L leucine, P proline, T threonine,[46]
- F PHenylalanine and R aRginine are assigned by being phonetically suggestive,[46]
- W tryptophane is assigned based on the double ring being visually suggestive to the bulky letter W,[46]
- K lysine and Y tyrosine are assigned as alphabetically nearest to their initials L and T (note that U was avoided for its similarity with V, while X was reserved for undetermined or atypical amino acids); for tyrosine the mnemonic tYrosine was also proposed,[47]
- D aspartate was assigned arbitrarily, with the proposed mnemonic asparDic acid;[48] E glutamate was assigned in alphabetical sequence being larger by merely one methylene –CH2– group,[47]
- N asparagine was assigned arbitrarily, with the proposed mnemonic asparagiNe;[48] Q glutamine was assigned in alphabetical sequence of those still available (note again that O was avoided due to similarity with D), with the proposed mnemonic Qlutamine.[48]

